Abstract
Background: The somatic KIT D816V mutation leads to an activation of the receptor tyrosine kinase KIT and represents a diagnostic criterion for systemic mastocytosis (SM). In the majority of patients, only a very few KIT D816V+ mast cells (MC) or MC precursors, if any, are found in peripheral blood (PB) and bone marrow (BM) aspirates. In contrast, the MC count in BM biopsies is typically much higher. Melting curve analysis after peptide nucleic acid-mediated PCR clamping (clamp-PCR) is widely used for detecting KIT mutations in biopsies.1 We have previously proposed dPCR as a new standard method of KIT D816V testing in SM that can also reliably quantify the variant allele fraction (VAF) in formalin-fixed paraffin-embedded (FFPE) material.2 While multilineage involvement of KIT D816V indicated by a high VAF in PB was associated with an aggressive clinical course,3-6 the mutant allele burden in tissue sections has not been studied in a large series of SM patients.
Methods: The VAF of KIT D816V in the tissue was assessed by dPCR (PrimePCR ddPCR, Biorad) on DNA isolated from 211 FFPE BM sections from 116 patients with SM (median age 53 years; 58 females, 58 males) and 57 controls (lymphoma patients undergoing staging biopsy). A total of 91 SM patients were diagnosed with indolent SM (ISM) and 25 with advanced SM (aggressive SM, SM with associated hematologic neoplasm, and mast cell leukemia according to WHO criteria). Results of diagnostic samples were compared to qualitative analysis of KIT D816V clamp-PCR. BM MC infiltration was quantified in a representative area of the tryptase stained section and expressed as % of nucleated cells. Total tryptase levels in serum were determined by a fluoroenzyme-immunoassay.
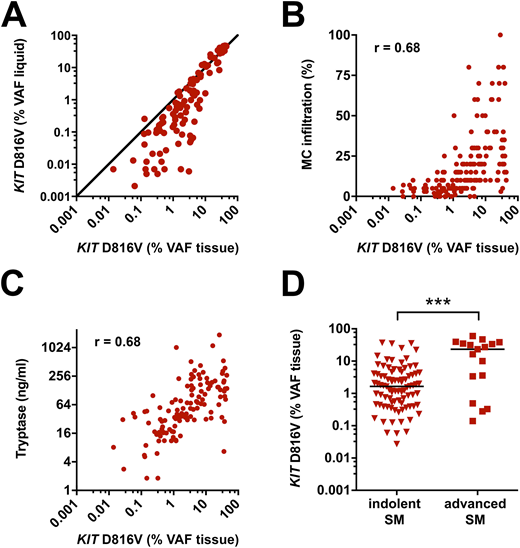
Results: The KIT D816V mutation burden in FFPE BM sections of SM patients showed marked differences between patients ranging from 0.027% to 60% VAF (median: 1.9%). The VAF in BM sections was largely independent from that in liquid specimen (PB and BM aspirate). In particular, a number of ISM patients showed substantially higher VAF in FFPE tissue than in liquid specimen (Figure 1A). In line with this, a higher correlation of KIT D816V mutation burden to BM MC infiltration and serum tryptase levels was observed for FFPE tissue (Spearman's coefficient of correlation r = 0.68 and r = 0.68 respectively; Figure 1B-C) than for liquid specimen (r = 0.48 and r = 0.58 respectively). When analyzing subgroups of SM, FFPE BM sections of patients with advanced SM had a significantly higher KIT D816V allele burden (median 23.40%) compared to patients with ISM (median 1.65%; p<0.001 Figure 1D). Finally, we assessed the sensitivity and specificity of dPCR for KIT D816V analysis in FFPE BM section compared to clamp PCR. While a specificity of 100% was observed for both assays, dPCR showed a significantly higher sensitivity to detect the mutation in BM tissue of ISM patients compared to clamp PCR (97% vs. 89%; p<0.05).
Conclusion: dPCR is a sensitive method to detect KIT D816V and reliably quantify the mutant allele burden in FFPE BM sections of SM patients. The mutational burden in the tissue represents a new molecular marker of the disease burden in SM since it reflects both the multilineage involvement of KIT D816V and the MC burden while the mutation burden in PB primarily reflects the KIT D816V multilineage involvement. Although the tissue mutational burden in patients with advanced SM is higher compared to ISM, the difference is not as substantial as described for PB. Thus, the prognostic value of KIT D816V allele burden measurement in SM needs to be redefined for BM sections. Potential future applications of KIT D816V quantification in FFPE BM sections include treatment response monitoring and definition of high disease burden in smoldering SM (SSM). We propose to include dPCR-based KIT D816V mutant allele burden measurement in BM tissue as a new biomarker for disease burden in SM in future clinical trials.
References:
1. Sotlar K, et al. Am J Pathol. 2003;162(3):737-746.
2. Greiner G, et al. Clin Chem. 2018;64(3):547-555.
3. Erben P, et al. Ann Hematol. 2014;93(1):81-88.
4. Hoermann G, et al. Allergy. 2014;69(6):810-813.
5. Jara-Acevedo M, et al. Mod Pathol. 2015;28(8):1138-1149.
6. Broesby-Olsen S, et al. J Allergy Clin Immunol. 2013;132(3):723-728.
Sperr:Novartis: Honoraria; Pfizer: Honoraria; Daiichi Sankyo: Honoraria. Valent:Incyte: Honoraria; Pfizer: Honoraria; Novartis: Honoraria. Hoermann:Novartis: Honoraria, Research Funding; Bristol-Myers Squibb: Honoraria; Pfizer: Honoraria.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal